COMP 5329
Lecture 2
P9 XOR
P31 Activation
P36 CE
P37 KL, Entropy
P38 SoftMax
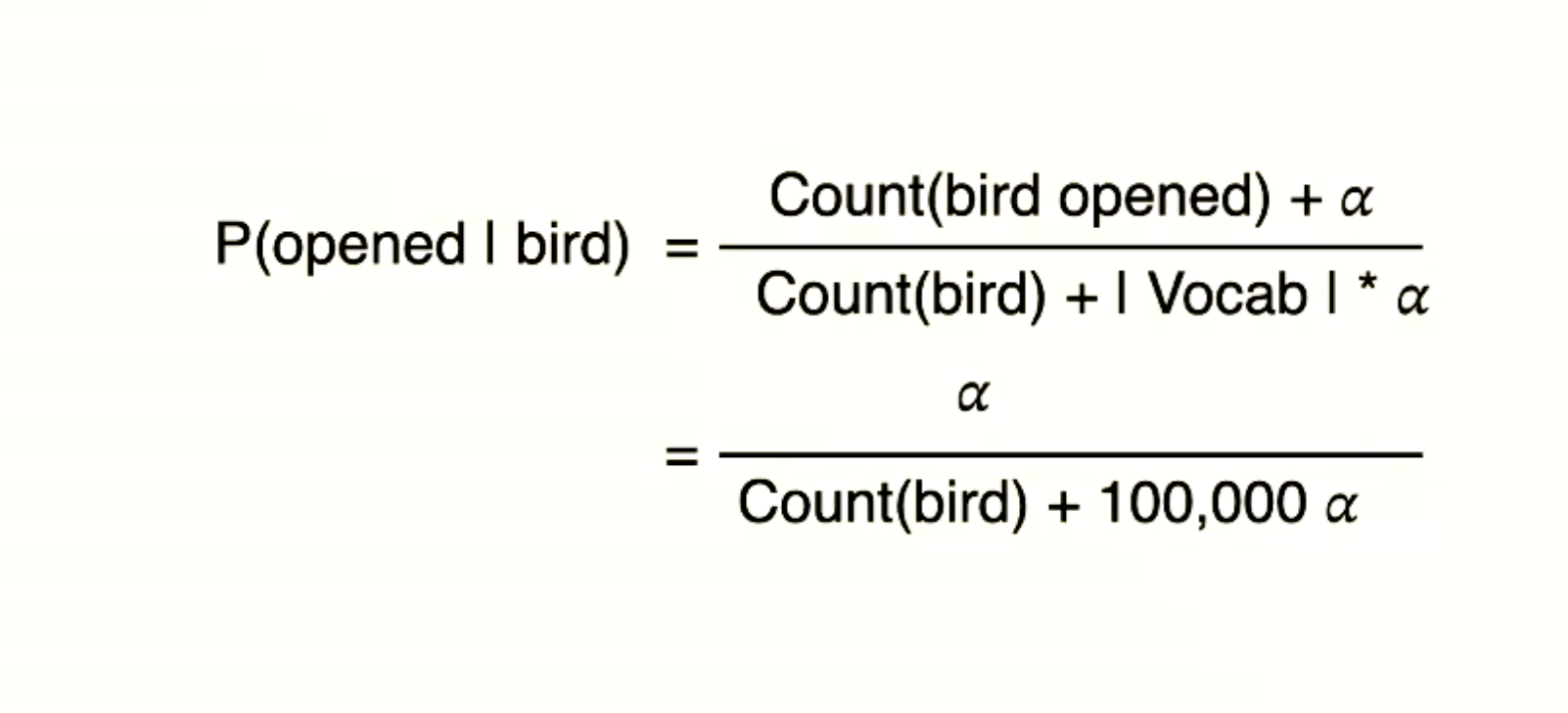
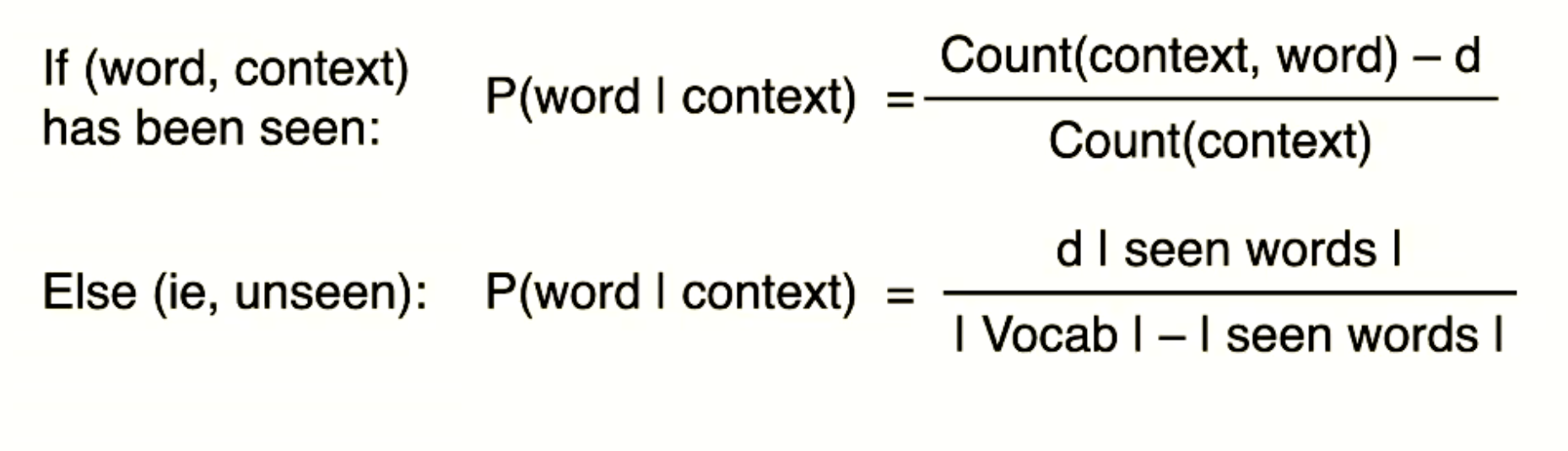
p36, p37 two attributes cross-entropy loss issue?
the disadvantage of batch gradient descent is when N is too large, the computation is very expensive, but the one example SGD only uses the single example. It may not be the best, due to the random.
mini-batches SGD: divide into mini-batches, in each epoch calculate the gradient
extension content:
sensitivity? make the back propagation similar to the feedforward
Lecture 3 GD
Different gradient descent.
Challenges: proper lr, same lr, saddle point
P9-10 Momentum
P11 NAG
P17 Adagrad
P21 Adadelta
P25 RMSprop
P26, 27 Adam
P34 Initialization
Batch Gradient descent: Accuracy
Stochastic gradient descent: efficiency
Mini-batch gradient descent: trade-off of accuracy and efficiency
Momentum is to update SGD, to accelerate and dampen oscillation
increase the momentum term in the same direction, reduce in change direction
NAG:
The big jump in the standard momentum is too aggressive
So, in the Nesterov accelerated gradient use a previous gradient to big jump and a correction
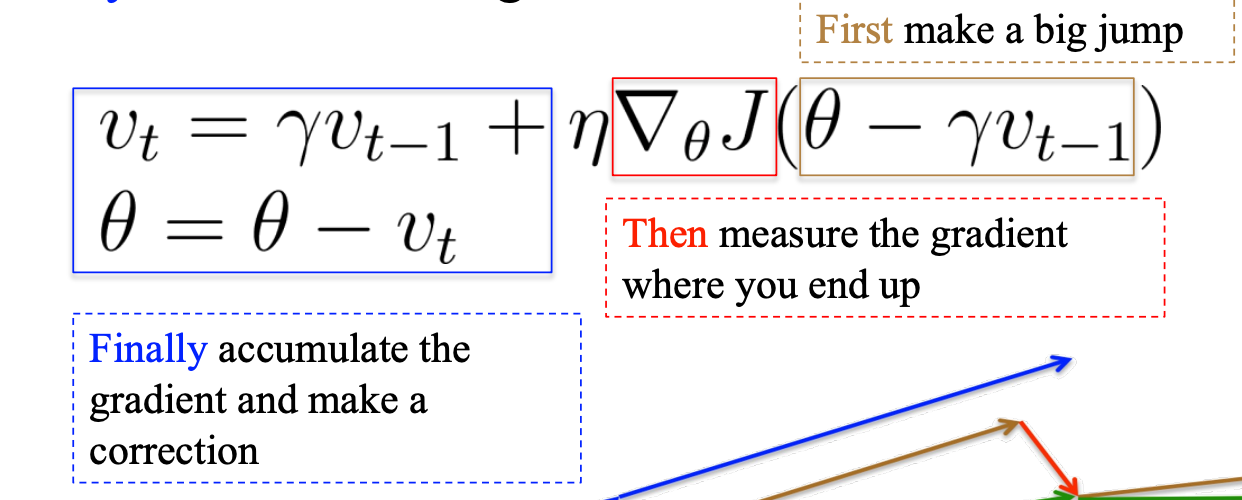
theta - r v_t-1 means that update according to the previous, which is the big jump depending on the previous accumulated gradient.
Then, calculate the gradient plus the contributed previously accumulated gradient to update the theta
Adagrad:
to achieve the different learning rate for features
i means dimension
t means iteration
suitable for sparse data
but the global learning rate needed
Adadelta:

in order to deal with the problem of Adagrad(infinitesimally small for the denominator in the end), modify the gt equation as above. Contributed the last square sum of gt, and current gt.
H is the secondary gradient
with consistent units
Lecture 4 Norm
P10 weight decay
pls write the structure of the normalization
inverted, dropout-connect
BN, (reduce covariance)
R means regularisation functions, r on page 6 is the upper boundary for parameter complex
Page 24 in slide, bottom 2 are independent methods
the group right top corner is the normal training process(i.e. only train on the training set)
drop out
scale down
in the training process, each unit has with p possibility
p present
1-p disappear
in the test process, they always present
p is times the w in layer
inverted
see slide
M in slide 36 is the matrix of binary (1 or 0)
adding the extra linear function after the normalization in each layer’s output. gamma means the scale parameter, beta means the shift parameter
batch norm: norm applies in all examples in this batch in each channel
layer norm: norm applies in each data sample in this batch
instance norm: norm applies in an example in this batch in a channel
group norm: norms apply in some channels in an example (split an example into multiple parts) in a data batch
Lecture 5 CNN
P46 different sorts of pooling
Spectral pooling?
un-pooling uses the (max) location information to reverse the process, keeping the 0 at information lacking place
more previous layer, the information is more simple. (line, color block)
more higher layer, the more meaningful information included
transposed convolution is also called the deconvolution method, enlarging the feature map until keeps the same as input size
in the process of deconvolution, the output will larger, it will happen to overlap in the process, summation the overlap region. Also, it has the stride, crop (like the padding, but crop the data in the output) Output size = (N-1) S + K - 2C
In pytorch, using the sequential in init rather than init function like tut note.
the kernel in CNN is similar to the digital, tut 6 notebook
its like the mouse in 5318, get a bigger score of the kernel output
Lecture 7
dead relu means that the zero output of the relu activation function
Data augmentation is rotate, distortion, color changing etc. methods to manipulate the data.
In local response normalization, a is the center of the region
overlapping pooling, polling stride less than pooling kernel size
smaller filter means deeper, more non-linearity, fewer parameters.
p is changing to ensure the size of the output is same on the googlenet
p28, right is the google net structure, and left is the normal way. It helps to decrease the depth of the feature map in the right method.
p37 is used to avoid the overconfident of the model, with the label smoothing
1 by 1 conv layer is used to exchange the information among channels ????
Shuffle net is another way to exchange information
Extension
Ghost filter?
Lego filter?
adder filter?
Lecture 8
Lecture 9
The masked LM in BERT in used to predict the masked word by linear regression
Classification in BERT is used in the binary classifier to identify the 1 or 2 sentences
P49 the place of dot product is, output of the classifier and the representation of token, multiple numbers is because the multiple-classifier of the model, then using the result passing into the softmax to predict the final prediction
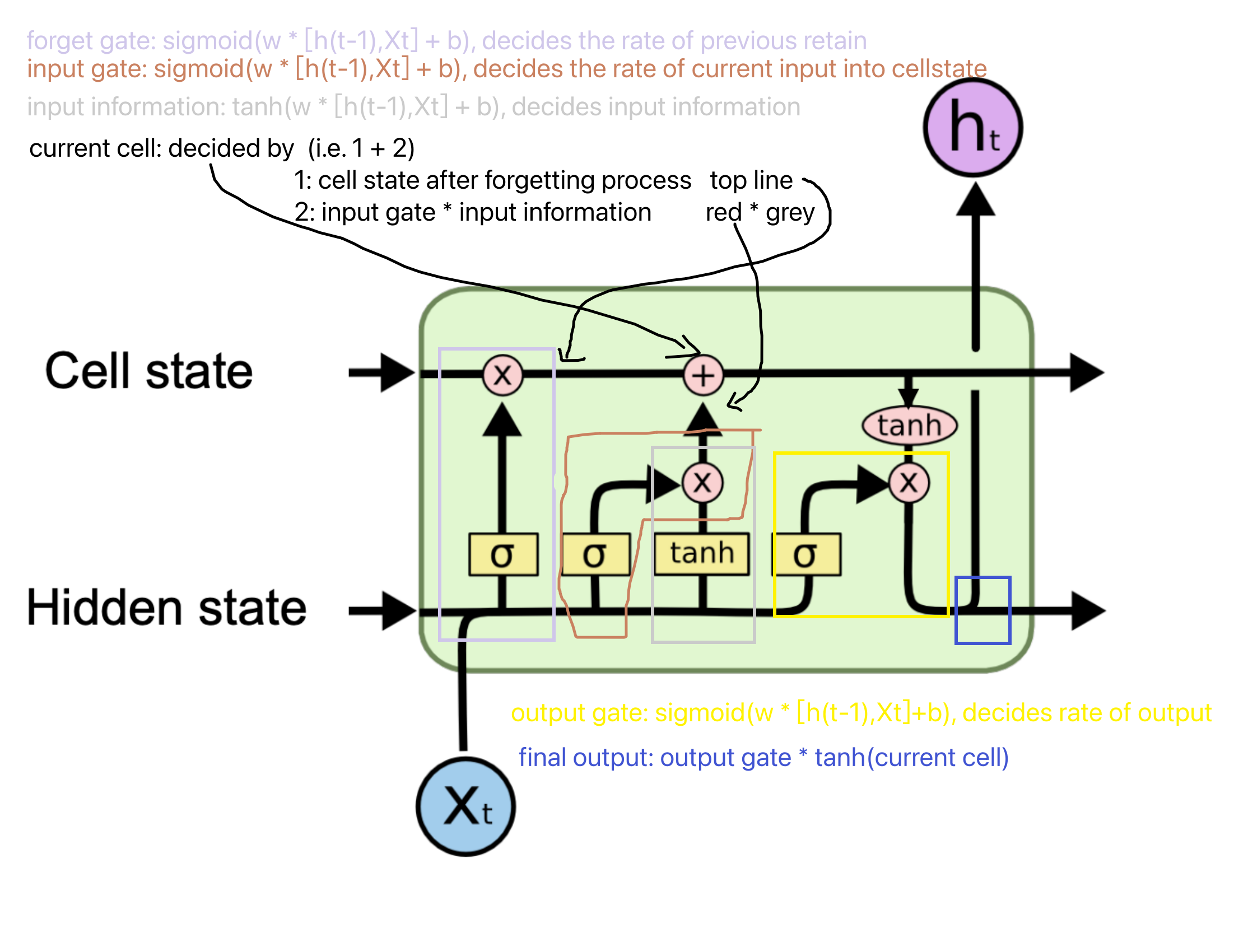
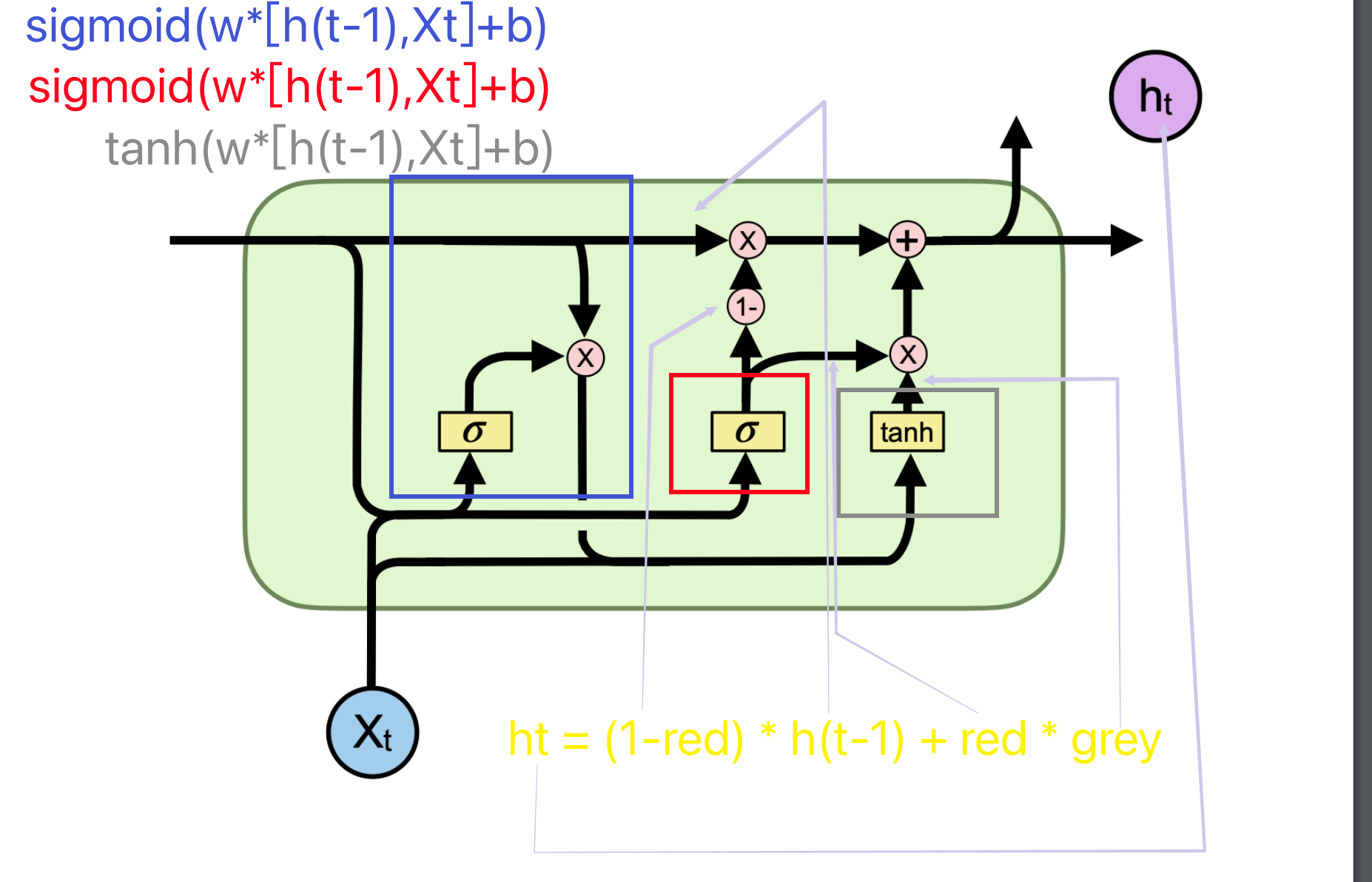
the gates in code split the matrix into multiple chunks and pass into the gates.
Lecture 10
P26: if the degree of the J is large, the information from J to I node is very small. Because the denominator is Djj Dii.
the D^-0.5 A D^-0.5(connection) H(value of each neighbour, near features) W(parameter)
Laplacian means to reduce the shape of the matrix, in the spectral matrix, the shape is quality large
Each row and each column in matrix of Laplacian are zero

the eigen matrix are used to reshape the original matrix, which similar to the PCA, focusing on the important part of the matrix
Lecture 11
the detected size of the boundary box many different. So, the resizing of the image is necessary for CNN. P16.
The proposal may overlap, it makes the duplication of calculation. P17
Fast R-CNN: instead of using the image level input like P16, the fast way is using the features in CNN as input to the bounding box task. P20
But the problem of different sizes still exists, fast R-CNN uses the max pooling way to ensure the size of the output.
Faster R-CNN: spp-net, improve at the extra pooling layer then fast R-CNN. In this model, the classification, detection, boundary box, etc tasks are all done by different CNNs.
In other words, the nature of the faster here is replacing the original image object detection by the feature object detection. It reduces the computational resources.
Mask R-CNN: deter the pixel in the image along to what label.
RoIAlign is more accurate in the pooling layer, directly dividing the result evenly without considering the number, and using the distance of each point to decide the number of the results after the max pooling.
excepted predict the xyhw, the dx,dy,dh,dw (difference of the xyhw) are also included in the prediction. Jargon called an anchor.
Lecture 12
high value in Dx, maximize the (1-D(G(z)))
Lecture 13
diffusion
GAN: 输入为随机输入
f(theta) = Y
Y 为图像
X 为随机输入
F()为神经网络, 映射xy之间的转换关系,从正态分布到图像的分布(特殊的集合)
不用MSE的原因是 噪音和图像之前的关系没有有意义的关系,也就是梯度会没有, 并且只能生成看到过的图片, 不会生成没看见过的图片
生成式对抗模型:两个神经网络,生成式网络(生成假图)和分类器(检验真假)
生成式模型从分类起学会到如何生成真图
分类无法区分的时候,就训练完了
VAE: encoder the image, then decode it
decode之后和原始的 计算loss
从参化
diffusion: 去燥模型,退热定律
X → XT.noisy(高斯)
X → X+Theta → XT.noisy(高斯)
X → … → X+Theta * n → XT.noisy(高斯)
每次加一点点噪音, 直到纯噪音
每次加的高斯分布的噪音
loss 为检测图片中的噪音数量
反过去,就是按照噪音去预测图片
diffusion只有一个NN,其他模型为多个